Introduction: Despite the wider use of midostaurin (MIDO) in combination with intensive chemotherapy (ICT) as the 1st-line treatment for FLT3-mutated acute myeloid leukemia (AML), complete remission (CR) rates are close to 60%, and relapses occur in over 40% of cases, demonstrating the ability of leukemic cells to resist and evade therapy ( Stone et al., NEJM 2017). Conventional fragment-length analyses of paired diagnosis/relapse samples have shown that FLT3-internal tandem duplications (ITDs) are retained in 80% and 50% of cases following ICT alone and MIDO+ICT respectively ( Schmalbrock et al., Blood 2021). Only limited data are available on the dynamics of FLT3-ITDs or other co-mutations in refractory patients (pts). Here, we conducted a retrospective study involving 115 pts with relapsed/refractory AML harboring FLT3-ITD at diagnosis.
Materials and methods: Clonal evolution was examined in paired diagnosis/progression blood or bone marrow samples from 115 pts with FLT3-ITD+ AML treated with MIDO+ICT (n=33) or ICT alone (n=82). Among them, 21 pts had primary refractory disease (MIDO+ICT, n=8; ICT, n=13) and 94 pts relapsed after achieving CR (MIDO+ICT, n=25; ICT, n=69). FLT3-ITDs and co-mutations were screened on genomic DNA by high-throughput sequencing at both timepoints using a custom-designed panel. For accurate annotation and quantification of FLT3-ITDs from sequencing data, we used the recentlypublished FiLT3r algorithm ( Boudry et al., BMC Bioinformatics 2022). For each ITD detected, FiLT3r allelic ratio (AR) was assessed by the ratio between the mutated allele and the wild-type allele.
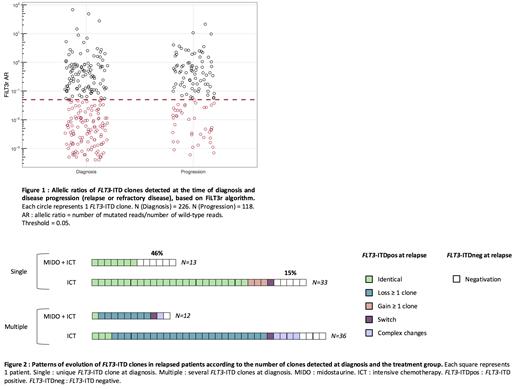
Results: A total of 226 FLT3-ITDs were detected in 115 pts at AML diagnosis, among which 120 (53%) ITDs were found with an AR below 0.05 ( Figure 1).
Among pts who achieved CR and experienced relapse (n=94), 48 had multiple FLT3-ITDs at diagnosis and 46 had a single FLT3-ITD at diagnosis. Overall, we observed a simplification of the FLT3-ITD repertoire upon relapse with the persistence of at least one initial clone in 8/12 [67%] and 24/36 [67%] pts with multiple ITDs receiving MIDO+ICT and ICT alone respectively. In relapsed pts who initially had a single FLT3-ITD clone at diagnosis, the addition of MIDO to ICT was associated with a higher rate of FLT3-ITD negativity compared to pts receiving ICT alone (6/13 [46%] vs 5/33 [15%]; P = 0.05) ( Figure 2).
Interestingly, among 21 pts with primary refractory AML, we observed that FLT3-ITD mutation status became negative in 5/8 pts (62%) and 2/13 pts (15%) after induction with MIDO+ICT and ICT alone respectively.
We then compared the initial characteristics between retained and lost FLT3-ITDs at AML relapse. Lost FLT3-ITDs had significantly lower AR than retained clones in both treatment groups. In order to limit the impact of sample dilution on the allele burden, we defined adjusted variant allele frequencies (VAFs) as the VAFs of FLT3-ITDs normalized to the VAFs of NPM1 mutations, whenever applicable. In so doing, we observed that adjusted VAFs of retained FLT3-ITDs increased at relapse, regardless of the treatment group (adjusted VAF, diagnosis vs relapse: 0.28 vs 0.86 and 0.88 vs 1.6 in the MIDO+ICT group and ICT alone group; P = 2.3e-03 and P = 2.4e-04). Importantly, an adjusted VAF higher than 1 was strongly suggestive of a homozygous state of FLT3-ITD. Such situation was found to be more prevalent at relapse in both treatment groups. Besides the selection of a dominant FLT3-ITD clone, other relapse-related changes including the acquisition of additional mutations will be presented.
Conclusion: Our study of the clonal dynamics of AML with FLT3-ITD mutations provides insights into the mechanisms underlying therapy escape. Our data suggest that clonal interference characterized by multiple FLT3-ITD clones is associated with a greater ability to select a FLT3-ITD-positive clone at relapse in pts receiving MIDO+ICT. Although the addition of MIDO to ICT increases the probability of eradicating a single FLT3-ITD clone, FLT3-ITD+ relapses remain common following this combination, often with the selection of homozygous FLT3-ITD clones and/or the emergence of new mutations. Finally, our data in refractory situation emphasize the need to reassess mutational status at each stage of progression before implementing targeted therapy.
Disclosures
Dombret:Jazz Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees, Research Funding; Pfizer: Research Funding; Servier: Membership on an entity's Board of Directors or advisory committees, Research Funding; Astellas: Research Funding; Incyte: Membership on an entity's Board of Directors or advisory committees. Pigneux:Jazz Pharmaceuticals: Honoraria, Membership on an entity's Board of Directors or advisory committees; Gilead: Honoraria; Abbvie: Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Support for attending meetings; Servier: Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Support for attending meetings, Research Funding; Roche: Research Funding; BMS: Membership on an entity's Board of Directors or advisory committees, Research Funding; Astellas: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Honoraria; Pfizer: Membership on an entity's Board of Directors or advisory committees. Recher:Jazz Pharmaceuticals: Other: Personal fees, Research Funding; Novartis: Other: Personal fees; Astellas: Other: Personal fees; BMS: Other: Personal fees, Research Funding; Amgen: Research Funding; Abbvie: Honoraria; Servier: Other: Personal fees; MaatPharma: Research Funding; IQVIA: Research Funding; Takeda: Other: Personal fees. Dumas:Novartis: Honoraria, Other: Research support for institution; Servier: Honoraria, Other: Research support for institution; BMS: Honoraria, Other: Research support for institution; Abbvie: Honoraria; Astellas: Honoraria, Other: Research support for institution; Daiichi-Sankyo: Honoraria, Other: Research support for institution; Jazz pharmaceutical: Honoraria; Janssen: Honoraria; Roche: Other: Research support for institution.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal